MUTATE: IN SILICO PERFORMANCE OF THE FLUCTUATION TEST
Hints
- After download Mutate requires the Java 5.0 (or higher) Runtime Environment freely available at http://www.java.com.
- Whatever the operating system, you should be able to run the program just by double clicking on the mutate.jar file
- WINDOWS USERS: If double-clicking the .jar file does not open the program, make sure that the jar extension is associated with javaw.exe (in your JRE folder).
Contact
For any questions you can contact me
Return to AC-R home
Sequences
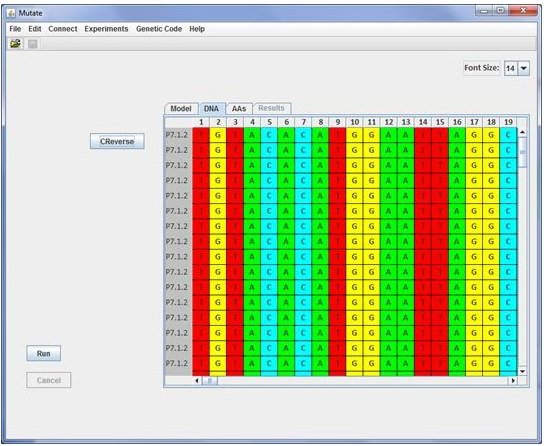
Before performing the fluctuation test the user should load a DNA sequence file which will be shown in the screen of the DNA panel.
The sequence format should be Fasta or Phylip sequential. The DNA panel is useful because it gives to the student the opportunity of
searching and managing sequences while reviewing concepts as genetic codes, reading frame and different kind of mutations.
For example, in the Experiments settings menu the user can define the type of mutation that generates resistance.
Therefore if in the Mutations checkbox, only STOP type is selected then the student is obligated to introduce such kind of mutation
before performing the fluctuation test. The expected number of mutations will be kÁNt where Á is the per site mutaiton rate and
k is the number of
mutations of the user-defined type (STOP codons in our example). The mutation can be introduced in any sequence.
However only one mutation per site is counted for contributing to the k add up which means that if we have ten sequences and mutate
the same site in each, say position 7, the value of k will be just 1. On the contrary, if positions 7 and 8 are mutated the value of k
will be 2 in any case.
A. Carvajal-Rodriguez - Departamento de Bioquímica Genética e Inmunología - Universidad de Vigo.
( Last update: December 2011)